Advanced Jupyter Notebooks
Tutorial: Advanced Jupyter Notebooks

Lying at the heart of modern data science and analysis is the Jupyter project lifecycle. Whether you’re rapidly prototyping ideas, demonstrating your work, or producing fully fledged reports, notebooks can provide an efficient edge over IDEs or traditional desktop applications.
Following on from Jupyter Notebook for Beginners: A Tutorial, this guide will be a Jupyter Notebooks tutorial that takes you on a journey from the truly vanilla to the downright dangerous. That’s right! Jupyter’s wacky world of out-of-order execution has the power to faze, and when it comes to running notebooks inside notebooks, things can get complicated fast.
This Jupyter Notebooks tutorial aims to straighten out some sources of confusion and spread ideas that pique your interest and spark your imagination. There are already plenty of great listicles of neat tips and tricks, so here we will take a more thorough look at Jupyter’s offerings.
This will involve:
Warming up with the basics of shell commands and some handy magics, including a look at debugging, timing, and executing multiple languages.
Exploring topics like logging, macros, running external code, and Jupyter extensions.
Seeing how to enhance charts with Seaborn, beautify notebooks with themes and CSS, and customise notebook output.
Finishing off with a deep look at topics like scripted execution, automated reporting pipelines, and working with databases.
If you’re a JupyterLab fan, you’ll be pleased to hear that 99% of this is still applicable and the only difference is that some Jupyter Notebook extensions aren’t compatible with JuputerLab. Fortunately, awesome alternatives are already cropping up on GitHub.
Now we’re ready to become Jupyter wizards!
Shell Commands
Every user will benefit at least from time-to-time from the ability to interact directly with the operating system from within their notebook. Any line in a code cell that you begin with an exclamation mark will be executed as a shell command. This can be useful when dealing with datasets or other files, and managing your Python packages. As a simple illustration:
It is also possible to use Python variables in your shell commands by prepending a $ symbol consistent with bash style variable names.
Note that the shell in which ! commands are executed is discarded after execution completes, so commands like cd will have no effect. However, IPython magics offer a solution.
Basic Magics
Magics are handy commands built into the IPython kernel that make it easier to perform particular tasks. Although they often resemble unix commands, under the hood they are all implemented in Python. There exist far more magics than it would make sense to cover here, but it’s worth highlighting a variety of examples. We will start with a few basics before moving on to more interesting cases.
There are two categories of magic: line magics and cell magics. Respectively, they act on a single line or can be spread across multiple lines or entire cells. To see the available magics, you can do the following:
As you can see, there are loads! Most are listed in the official documentation, which is intended as a reference but can be somewhat obtuse in places. Line magics start with a percent character %, and cell magics start with two, %%.
It’s worth noting that ! is really just a fancy magic syntax for shell commands, and as you may have noticed IPython provides magics in place of those shell commands that alter the state of the shell and are thus lost by !. Examples include %cd, %alias and %env.
Let’s go through some more examples.
Autosaving
First up, the %autosave magic let’s you change how often your notebook will autosave to its checkpoint file.
It’s that easy!
Displaying Matplotlib Plots
One of the most common line magics for data scientists is surely %matplotlib, which is of course for use with the most popular plotting libary for Python, Matplotlib.
Providing the inline argument instructs IPython to show Matplotlib plot images inline, within your cell outputs, enabling you to include charts inside your notebooks. Be sure to include this magic before you import Matplotlib, as it may not work if you do not; many import it at the start of their notebook, in the first code cell.
Now, let’s start looking at some more complex features.
Debugging
The more experienced reader may have had concerns over the ultimate efficacy of Jupyter Notebooks without access to a debugger. But fear not! The IPython kernel has its own interface to the Python debugger, pdb, and several options for debugging with it in your notebooks. Executing the %pdb line magic will toggle on/off the automatic triggering of pdb on error across all cells in your notebook.
This exposes an interactive mode in which you can use the pdb commands.
Another handy debugging magic is %debug, which you can execute after an exception has been raised to delve back into the call stack at the time of failure.
As an aside, also note how the traceback above demonstrates how magics are translated directly into Python commands, where %pdb became get_ipython().run_line_magic('pdb', ''). Executing this instead is identical to executing %pdb.
Timing Execution
Sometimes in research, it is important to provide runtime comparisons for competing approaches. IPython provides the two timing magics %time and %timeit, which each has both line and cell modes. The former simply times either the execution of a single statement or cell, depending on whether it is used in line or cell mode.
And in cell mode:
The notable difference of %timeit from %time is that it runs the specified code many times and computes an average. You can specify the number of runs with the -n option, but if nothing is passed a fitting value will be chosen based on computation time.
Executing Different Languages
In the output of %lsmagic above, you may have noticed a number of cell magics named after various programming, scripting or markup langauges, including HTML, JavaScript, Ruby, and LaTeX. Using these will execute the cell using the specified language. There are also extensions available for other languages such as R.
For example, to render HTML in your notebook:
This is really neat!
Similarly, LaTeX is a markup language for displaying mathematical expressions, and can be used directly:
Some important equations:
Configuring Logging
Did you know that Jupyter has a built-in way to prominently display custom error messages above cell output? This can be handy for ensuring that errors and warnings about things like invalid inputs or parameterisations are hard to miss for anyone who might be using your notebooks. An easy, customisable way to hook into this is via the standard Python logging module.
(Note: Just for this section, we’ll use some screenshots so that we can see how these errors look in a real notebook.)

The logging output is displayed separately from print statements or standard cell output, appearing above all of this.

This actually works because Jupyter notebooks listen to both standard output streams, stdout and stderr, but handle each differently; print statements and cell output route to stdout and by default logging has been configured to stream over stderr.
This means we can configure logging to display other kinds of messages over stderr too.

We can customise the format of these messages like so:
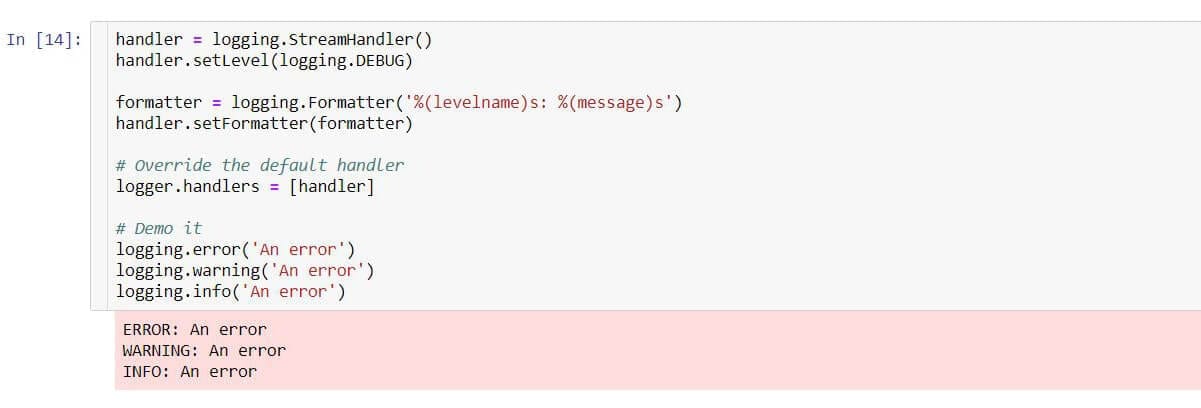
Note that every time you run a cell that adds a new stream handler via logger.addHandler(handler), you will receive an additional line of output each time for each message logged. We could place all the logging config in its own cell near the top of our notebook and leave it be or, as we have done here, brute force replace all existing handlers on the logger. We had to do that in this case anyway to remove the default handler.
It’s also easy to log to an external file, which might come in handy if you’re executing your notebooks from the command line as discussed later. Just use a FileHandler instead of a StreamHandler:
A final thing to note is that the logging described here is not to be confused with using the %config magic to change the application’s logging level via %config Application.log_level="INFO", as this determines what Jupyter outputs to the terminal while it runs.
Extensions
As it is an open source webapp, plenty of extensions have been developed for Jupyter Notebooks, and there is a long official list. Indeed, in the Working with Databases section below we use the ipython-sql extension. Another of particular note is the bundle of extensions from Jupyter-contrib, which contains individual extensions for spell check, code folding and much more.
You can install and set this up from the command line like so:
This will install the jupyter_contrib_nbextensions package in Python, install it in Jupyter, and then enable the spell check and code folding extensions. Don’t forget to refresh any notebooks live at the time of installation to load in changes.
Note that Jupyter-contrib only works in regular Jupyter Notebooks, but there are new extensions for JupyterLab now being released on GitHub.
Enhancing Charts with Seaborn
One of the most common exercises Jupyter Notebook users undertake is producing plots. But Matplotlib, Python’s most popular charting library, isn’t renowned for attractive results despite it’s customisability. Seaborn instantly prettifies Matplotlib plots and even adds some additional features pertinent to data science, making your reports prettier and your job easier. It’s included in the default Anaconda installation or easily installed via pip install seaborn.
Let’s check out an example. First, we’ll import our libraries and load some data.
Seaborn provides some built-in sample datasets for documentation, testing and learning purposes, which we will make use of here. This “tips” dataset is a pandas DataFrame listing some billing information from a bar or restaurant. We can see the size of the total bill, the tip, the gender of the payer, and some other attributes.
total_bill
tip
sex
smoker
day
time
size
0
16.99
1.01
Female
No
Sun
Dinner
2
1
10.34
1.66
Male
No
Sun
Dinner
3
2
21.01
3.50
Male
No
Sun
Dinner
3
3
23.68
3.31
Male
No
Sun
Dinner
2
4
24.59
3.61
Female
No
Sun
Dinner
4
We can easily plot total_bill vs tip in Matplotlib.

Plotting in Seaborn is just as easy! Simply set a style and your Matplotlib plots will automatically be transformed.

What an improvement, and from only one import and a single extra line! Here, we used the darkgrid style, but Seaborn has a total of five built-in styles for you to play with: darkgrid, whitegrid, dark, white, and ticks.
But we don’t have to stop with styling: as Seaborn is closely integrated with pandas data structures, its own scatter plot function unlocks additional features.

Now we get default axis labels and an improved default marker for each data point. Seaborn can also automatically group by categories within your data to add another dimension to your plots. Let’s change the colour of our markers based on whether the group paying the bill were smokers or not.

That’s pretty neat! In fact, we can take this much further, but there’s simply too much detail to go into here. As a taster though, let’s colour by the size of the party paying the bill while also discriminating between smokers and non-smokers.

Hopefully it’s becoming clear why Seaborn describes itself as a “high-level interface for drawing attractive statistical graphics.”
Indeed, it’s high-level enough to, for example, provide one-liners for plotting data with a line of best fit (determined through linear regression), whereas Matplotlib relies on you to prepare the data yourself. But if all you need is more attractive plots, it’s remarkably customisable; for example, if you aren’t happy with the default themes, you can choose from a whole array of standard color palettes or define your own.
For more ways Seaborn allows you to visualise the structure of your data and the statistical relationships within it, check out their examples.
Macros
Like many users, you probably find yourself writing the same few tasks over and over again. Maybe there’s a bunch of packages you always need to import when starting a new notebook, a few statistics that you find yourself computing for every single dataset, or some standard charts that you’ve produced countless times?
Jupyter lets you save code snippets as executable macros for use across all your notebooks. Although executing unknown code isn’t necessarily going to be useful for anyone else trying to read or use your notebooks, it’s definitely a handy productivity boost while you’re prototyping, investigating, or just playing around.
Macros are just code, so they can contain variables that will have to be defined before execution. Let’s define one to use.
Now, to define a macro we first need some code to use.
We use the %macro and %store magics to set up a macro that’s reusable across all our notebooks. It’s common to begin macro names with a double underscore to distinguish them from other variables, like so:
The %macro magic takes a name and a cell number (the number in the square brackets to the left of the cell; in this case 23 as in In [23]), and we’ve also passed -q to make it less verbose. %store actually allows us to save any variable for use in other sessions; here, we pass the name of the macro we created so we can use it again after the kernel is shut down or in other notebooks. Run without any parameters, %store lists your saved items.
To load the macro from the store, we just run:
And to execute it, we merely need to run a cell that solely contains the macro name.
Let’s modify the variable we used in the macro.
When we run the macro now, our modified value is picked up.
This works because macros just execute the saved code in the scope of the cell; if name was undefined we’d get an error.
But macros are far from the only way to share code across notebooks.
Executing External Code
Not all code belongs in a Jupyter Notebook. Indeed, while it’s entirely possible to write statistical models or even entire multi-part projects in Jupyter notebooks, this code becomes messy, difficult to maintain, and unusable by others. Jupyter’s flexibility is no substitute for writing well-structured Python modules, which are trivially imported into your notebooks.
In general, when your quick notebook project starts to get more serious and you find yourself writing code that is reusable or can be logically grouped into a Python script or module, you should do it! Aside from the fact that you can import your own modules directly in Python, Jupyter also lets you %load and %run external scripts to support better organised, larger-scale projects and reusability.
Tasks such as importing the same set of packages over and over for every project project are a perfect candidate for the %load magic, which will load an external script into the cell in which it’s executed.
But enough talk already, let’s look at an example! If we create a file imports.py containing the following code:
We can load this simply by writing a one-line code cell, like so:
Executing this will replace the cell contents with the loaded file.
Now we can run the cell again to import all our modules and we’re ready to go.
The %run magic is similar, except it will execute the code and display any output, including Matplotlib plots. You can even execute entire notebooks this way, but remember that not all code truly belongs in a notebook. Let’s check out an example of this magic; consider a file containing the following short script.
When executed via %run, we get the following result.

If you wish to pass arguments to a script, simply list them explicitly after the filename %run my_file.py 0 "Hello, World!" or using variables %run $filename {arg0} {arg1}. Additionally, use the -p option to run the code through the Python profiler.
Scripted Execution
Although the foremost power of Jupyter Notebooks emanates from their interactive flow, it is also possible to run notebooks in a non-interactive mode. Executing notebooks from scripts or the command line provides a powerful way to produce automated reports or similar documents.
Jupyter offers a command line tool that can be used, in its simplest form, for file conversion and execution. As you are probably aware, notebooks can be converted to a number of formats, available from the UI under “File > Download As”, including HTML, PDF, Python script, and even LaTeX. This functionality is exposed on the command line through an API called nbconvert. It is also possible to execute notebooks within Python scripts, but this is already well documented and the examples below should be equally applicable.
It’s important to stress, similarly to %run, that while the ability to execute notebooks standalone makes it possible to write all manor of projects entirely within Jupyter notebooks, this is no substitute for breaking up code into standard Python modules and scripts as appropriate.
On the Command Line
It will become clear later how nbconvert empowers developers to create their own automated reporting pipelines, but first let’s look at some simple examples. The basic syntax is:
For example, to create a PDF, simply write:
This will take the currently saved static content of notebook.ipynb and create a new file called notebook.pdf. One caveat here is that to convert to PDF requires that you have pandoc (which comes with Anaconda) and LaTeX (which doesn’t) installed. Installation instructions depend on your operating system.
By default, nbconvert doesn’t execute your notebook code cells. But if you also wish to, you can specify the --execute flag.
A common snag arises from the fact that any error encountered running your notebook will halt execution. Fortunately, you can throw in the --allow-errors flag to instruct nbconvert to output the error message into the cell output instead.
Parameterization with Environment Variables
Scripted execution is particularly useful for notebooks that don’t always produce the same output, such as if you are processing data that change over time, either from files on disk or pulled down via an API. The resulting documents can easily be emailed to a list of subscribers or uploaded to Amazon S3 for users to download from your website, for example.
In such cases, it’s quite likely you may wish to parameterize your notebooks in order to run them with different initial values. The simplest way to achieve this is using environment variables, which you define before executing the notebook.
Let’s say we want to generate several reports for different dates; in the first cell of our notebook, we can pull this information from an environment variable, which we will name REPORT_DATE. The %env line magic makes it easy to assign the value of an environment variable to a Python variable.
Then, to run the notebook (on UNIX systems) we can do something like this:
As all environment variables are strings, we will have to parse them to get the data types we want. For example:
And we simply parse like so:
Parsing dates is definitely less intuitive than other common data types, but as usual there are several options in Python.
On Windows
If you’d like to set your environment variables and run your notebook in a single line on Windows, it isn’t quite as simple:
Keen readers will notice the lack of a space after defining A_STRING and A_DATE above. This is because trailing spaces are significant to the Windows set command, so while Python will successfully parse the integer and the float by first stripping whitespace, we have to be more careful with our strings.
Parameterization with Papermill
Using environment variables is fine for simple use-cases, but for anything more complex there are libraries that will let you pass parameters to your notebooks and execute them. With over 1000 stars on GitHub, probably the most popular is Papermill, which can be installed with pip install papermill.
Papermill injects a new cell into your notebook that instantiates the parameters you pass in, parsing numeric inputs for you. This means you can just use the variables without any extra set-up (though dates still need to be parsed). Optionally, you can create a cell in your notebook that defines your default parameter values by clicking “View > Cell Toolbar > Tags” and adding a “parameters” tag to the cell of your choice.
Our earlier example that produced an HTML document now becomes:
We specify each parameter with the -p option and use an intermediary notebook so as not to change the original. It is perfectly possible to overwrite the original example.ipynb file, but remember that Papermill will inject a parameter cell.
Now our notebook set-up is much simpler:
Our brief glance so far uncovers only the tip of the Papermill iceberg. The library can also execute and collect metrics across notebooks, summarise collections of notebooks, and it provides an API for storing data and Matplotlib plots for access in other scripts or notebooks. It’s all well documented in the GitHub readme, so there’s no need to reiterate here.
It should now be clear that, using this technique, it is possible to write shell or Python scripts that can batch produce multiple documents and be scheduled via tools like crontab to run automatically on a schedule. Powerful stuff!
Styling Notebooks
If you’re looking for a particular look-and-feel in your notebooks, you can create an external CSS file and load it with Python.
This works because IPython’s HTML objects are inserted directly into the cell output div as raw HTML. In fact, this is equivalent to writing an HTML cell:
To demonstrate that this works let’s use another HTML cell.
This text has a nice colour
Using HTML cells would be fine for one or two lines, but it will typically be cleaner to load an external file as we first saw.
If you would rather customise all your notebooks at once, you can write CSS straight into the ~/.jupyter/custom/custom.css file in your Jupyter config directory instead, though this will only work when running or converting notebooks on your own computer.
Indeed, all of the aforementioned techniques will also work in notebooks converted to HTML, but will not work in converted PDFs.
To explore your styling options, remember that as Jupyter is just a web app you can use your browser’s dev tools to inspect it while it’s running or delve into some exported HTML output. You will quickly find that it is well-structured: all cells are designated with the cell class, text and code cells are likewise respectively demarked with text_cell and code_cell, inputs and outputs are indicated with input and output, and so on.
There are also various different popular pre-designed themes for Jupyter Notebooks distributed on GitHub. The most popular is jupyterthemes, which is available via pip install jupyterthemes and it’s as simple as running jt -t monokai to set the “monokai” theme. If you’re looking to theme JupyterLab instead, there is a growing list of options popping up on GitHub too.
Hiding Cells
Although it’s bad practice to hide parts of your notebook that would aid other people’s understanding, some of your cells may not be important to the reader. For example, you might wish to hide a cell that adds CSS styling to your notebook or, if you wanted to hide your default and injected Papermill parameters, you could modify your nbconvert call like so:
In fact, this approach can be applied selectively to any tagged cells in your notebook, making the TagRemovePreprocessor configuration quite powerful. As an aside, there are also a host of other ways to hide cells in your notebooks.
Working with Databases
Databases are a data scientist’s bread and butter, so smoothing the interface between your databases and notebooks is going to be a real boon. Catherine Devlin‘s IPython SQL magic extension let’s you write SQL queries directly into code cells with minimal boilerplate as well as read the results straight into pandas DataFrames. First, go ahead and:
With the package installed, we start things off by executing the following magic in a code cell:
This loads the ipython-sql extension we just installed into our notebook. Let’s connect to a database!
Here, we just connected to a temporary in-memory database for the convenience of this example, but you’ll probably want to specify details appropriate to your database. Connection strings follow the SQLAlchemy standard:
Yours might look more like postgresql://scott:tiger@localhost/mydatabase, where driver is postgresql, username is scott, password is tiger, host is localhost and the database name is mydatabase.
Note that if you leave the connection string empty, the extension will try to use the DATABASE_URL environment variable; read more about how to customise this in the Scripted Execution section above.
Next, let’s quickly populate our database from the tips dataset from Seaborn we used earlier.
We can now execute queries on our database. Note that we can use a multiline cell magic %% for multiline SQL.
index
total_bill
tip
sex
smoker
day
time
size
0
16.99
1.01
Female
No
Sun
Dinner
2
1
10.34
1.66
Male
No
Sun
Dinner
3
2
21.01
3.5
Male
No
Sun
Dinner
3
You can parameterise your queries using locally scoped variables by prefixing them with a colon.
index
total_bill
tip
sex
smoker
day
time
size
0
16.99
1.01
Female
No
Sun
Dinner
2
1
10.34
1.66
Male
No
Sun
Dinner
3
2
21.01
3.5
Male
No
Sun
Dinner
3
The complexity of our queries is not limited by the extension, so we can easily write more expressive queries such as finding all the results with a total bill greater than the mean.
index
total_bill
tip
sex
smoker
day
time
size
0
2
21.01
3.50
Male
No
Sun
Dinner
3
1
3
23.68
3.31
Male
No
Sun
Dinner
2
2
4
24.59
3.61
Female
No
Sun
Dinner
4
And as you can see, converting to a pandas DataFrame was easy too, which makes plotting results from our queries a piece of cake. Let’s check out some 95% confidence intervals.

The ipython-sql extension also integrates with Matplotlib to let you call .plot(), .pie(), and .bar() straight on your query result, and can dump results direct to a CSV file via .csv(filename='my-file.csv'). Read more on the GitHub readme.
Wrapping Up
From the start of the Jupyter Notebooks tutorial for beginners through to here, we’ve covered a wide range of topics and really laid the foundations for what it takes to become a Jupyter master. These articles aim serve as a demonstration of the breadth of use-cases for Jupyter Notebooks and how to use them effectively. Hopefully, you have gained a few insights for your own projects!
There’s still a whole host of other things we can do with Jupyter notebooks that we haven’t covered, such as creating interactive controls and charts, or developing your own extensions, but let’s leave these for another day. Happy coding!
Reference : https://www.dataquest.io/blog/advanced-jupyter-notebooks-tutorial/
Last updated